Symbiota EML Export Module
The NEON Biorepository Data Portal is a Symbiota portal that curates not only biological specimens data, but also non-biological data, such as soil samples.
Biological specimens with reliable data are published in the GBIF network. However, there are also some collections that will never be published to GBIF for a variety of reasons.
To improve data discoverability, the NEON Biorepository Data Portal publishes all data that does not fit to GBIF into the Environmental Data Initiative (EDI) website.
While GBIF uses the Darwin Core standard, EDI uses the Ecological Metadata Language (EML) standard.
With this in mind, I was tasked with designing and coding a module that manipulates specimens’ data on demand to export data packages in the EML format (including Darwin Core compliant fields) ready to be published on the EDI Data Portal.
This module is available currently only at the NEON Biorepository Data Portal, but it can be adapted to other Symbiota portals if there is demand.
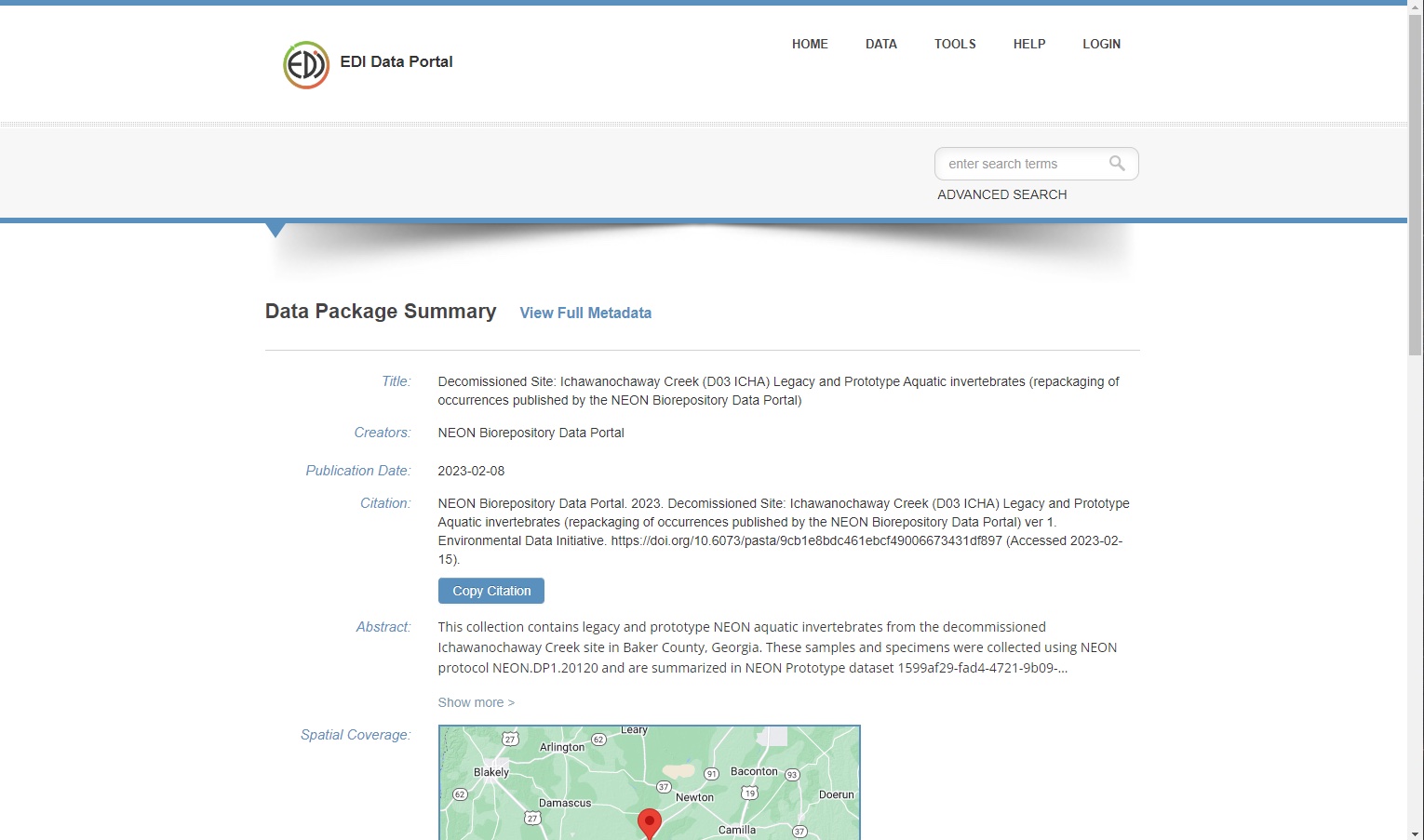 Screenshot of an example of a package produced by the EML export module at the NEON Biorepository Data Portal, and then published to EDI - See it on EDI
Screenshot of an example of a package produced by the EML export module at the NEON Biorepository Data Portal, and then published to EDI - See it on EDI
